※ User Guide
This website is free and open to all users and there is no login requirement.
Tutorial:
Frequently Asked Questions:
1.
Q: Is GPS-HALP useful?
A: 2.
Q: How to use GPS-HALP webserver?
A:
First, you could find the prediction website in "WEB SERVER" page of GPS-HALP. Second, enter protein sequence(s) in fasta format, which starts with a '>' followed with protein/peptide name. Then, select the His/Arg/Lys amino acid(s). Just click the "Submit" button, wait a moment, you can get the prediction results and annotations for H/R/K p-sites.
3.
Q: How to choose the different prediction modules? A:
We provide five modules of prediction for users. You can click "here >>" at "WEB SERVER" page to change the online service mode or just click the following link for species-specific prediction:
4.
Q: How to read the GPS-HALP results? A:
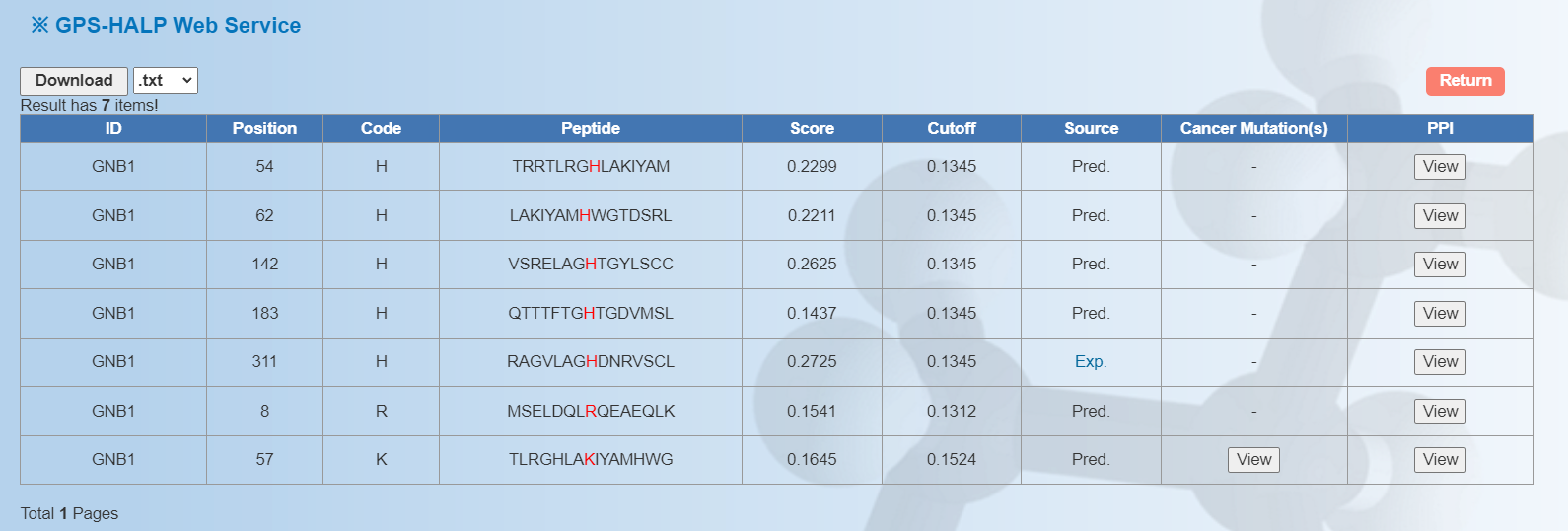
Here we use the human protein GNB1 as an example. After clicking "Submit", the prediction results of H,R and K phosphoamino sites with medium threshold are shown as follows:
ID: The name/id of the protein sequence that you input to predict. Position: The position of the site which is predicted to be phosphorylated. Code: The residue which is predicted to be phosphorylated. Peptide: The predicted phosphopeptide with 7 amino acids upstream and 7 amino acids downstream around the modified residue. Score: The value calculated by GPS-HALP algorithm to evaluate the potential of phosphorylation. The higher the value, the more potential the residue is phosphorylated. Cutoff: The cutoff value under the threshold. Different threshold means different precision, sensitivity and specificity. Source: Whether this phosphorylation site validated by experiment, "Exp." means YES, while "Pred." means NO. "Exp." links to the source site. Cancer Mutation(s): The mutation status for the potential phosphorylation site, integrated from TCGA [PMID: 29625055], ICGC [PMID: 30877282] and COSMIC [PMID: 30371878] databases. PPI: The protein-protein interaction status for the potential phosphorylation protein from BioGrid [PMID: 30476227]. Part 1: Part 2: Part 1: Part 2 : 5.
Q: How to choose the cut-off
values and the thresholds? A:
Firstly, we calculated the theoretically maximal
false positive rate (FPR) for each H/R/K p-sites predictors.
The three thresholds of GPS-HALP were decided based
on calculated FPRs. The high, medium and low thresholds were established
with FPRs of 2%, 6% and 10%. The same procedure was conducted for species-specific predictors.
6.
Q: What's the meaning of False
Positive Rate (FPR)? A:
The false positive rate (FPR) is the proportion of negative
sites that are erroneously predicted as positive hits.
Given a data set containing all of non-phosphorylation
sites, the real FPR could be easily computed. However,
precise calculation of FPR is unavailable due to lack
of a "gold-standard" negative data set. Here
we developed a simple and fast method to construct the
near-negative data set and estimate the theoretically
maximal FPRs. Firstly, we calculated the distribution
of amino acids composition in 29 species, including
S. cerevisiae, S. pombe, C. elegans, D. melanogaster, M. musculus,
and H. sapiens. Then we randomly generated
10,000 PSP(10,10) peptides to construct a near-negative
data set based on the real frequencies of twenty amino
acids from these species. Although there were a
few sites to be real hits, its proportion would be very
small. The process was repeated twenty times and the
average FPR was calculated by GPS-HALP as the theoretically
maximal FPR. Also, the negative sites could be randomly
retrieved from these species. And the results
from both methods are very similar. 7. Q: I have a few questions which
are not listed above, how can I contact the authors
of GPS-HALP? A:
Please contact the major author: Weizhi Zhang, Danyang Xu, and Dr.
Yu Xue for details. 8. Q:
Can I use GPS-HALP on different browsers?
A: Yes, we tested our web server on different browsers.
(1) The three non-canonical H/R/K phosphorylation sites (p-sites) are reported to have more and more important regulation roles in diverse biological processes. For example, phosphorylated histidine has been implied not only as a component of bacterial two-component signaling (TCS) systems but also involved in mammalian immune system generating phosphorylated histidine antibodies and further regulating cancer progression (Tony Hunter, 2022). In bacteria phosphorylated arginine was reported to play a role in the quality control of proteins (Débora Broch Trentini, et al,. 2016). The previous findings indicated that non-canonical phosphorylation could have functional significance across different species.
(2) GPS-HALP is a prediction tool to obtain three non-canonical H/R/K p-sites in silico. We validated that MAML technologies performed well on PTM predictions (Shui Ke, et al., 2023). We believe this tool would help other researchers focus on potential H/R/K p-sites which could be further verified in vitro/vivo.
(3) The web server of GPS-HALP could provide visualization of structure information for proteins containing potential H/R/K p-sites. Meanwhile, we annotated protein-protein interaction information, mutation information, tissue expression and immune cell expression for proteins with H/R/K p-sites. These information would accelerate users understanding of the multifaceted functions of potentially H/R/K phosphorylated proteins.
(1) GPS-HALP Web Server (ResNet): The default web server to provide H/R/K p-sites prediction and visualization function using ResNet model. We provide 3D structure, statistics, disorder propensity of protein and annotations from public resources.
(2) GPS-HALP Web Server (Integrated): Using integrated models to provide H/R/K p-sites prediction and visualization function. We provide 3D structure, statistics, disorder propensity of protein and annotations from public resources.
(3) GPS-HALP Web Server (Comprehensive Prediction): The comprehensive prediction appends calculation of secondary structure and surface accessibility.
(4) GPS-HALP Web Server (Species-specific Prediction): We provide 29 species for species-specific prediction.
(5) GPS-HALP Web Server (Prediction by Protein Identifiers): If you want to predict with gene name, protein name or UniProt Accession, please choose this one.
<1>. The table of the GPS-HALP results
<2>. The visualization of default prediction
Up: The visualization for the positional distribution of the predicted sites in protein sequence. By default, the 3 predicted p-sites in order of sequence are displayed.
Down: The visualization for protein disordered region predicted by IUPred
[PMID: 15955779]. Cutoff=0.5, if score of prediction > cutoff, the residue is considered in disordered region.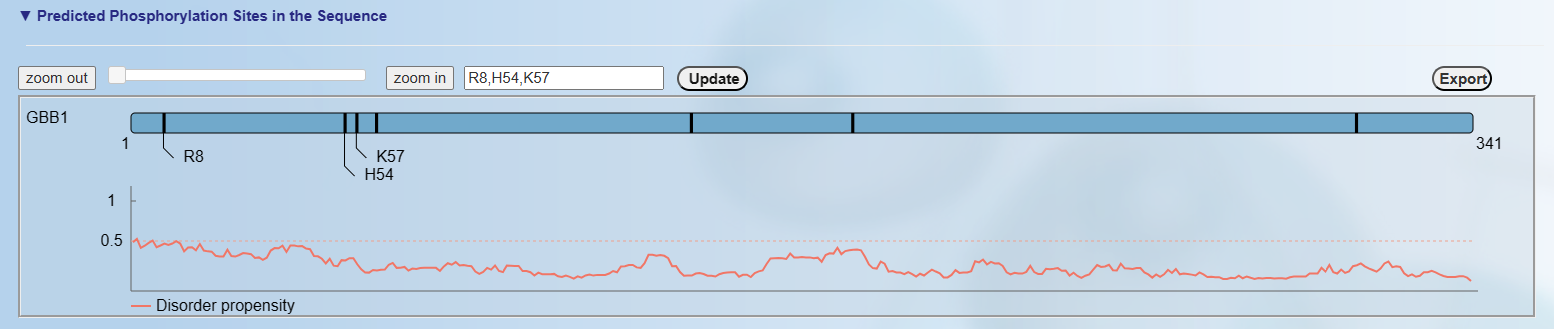
Upper left: The distribution of H/R/K sites and distribution of H/R/K sites in disordered region.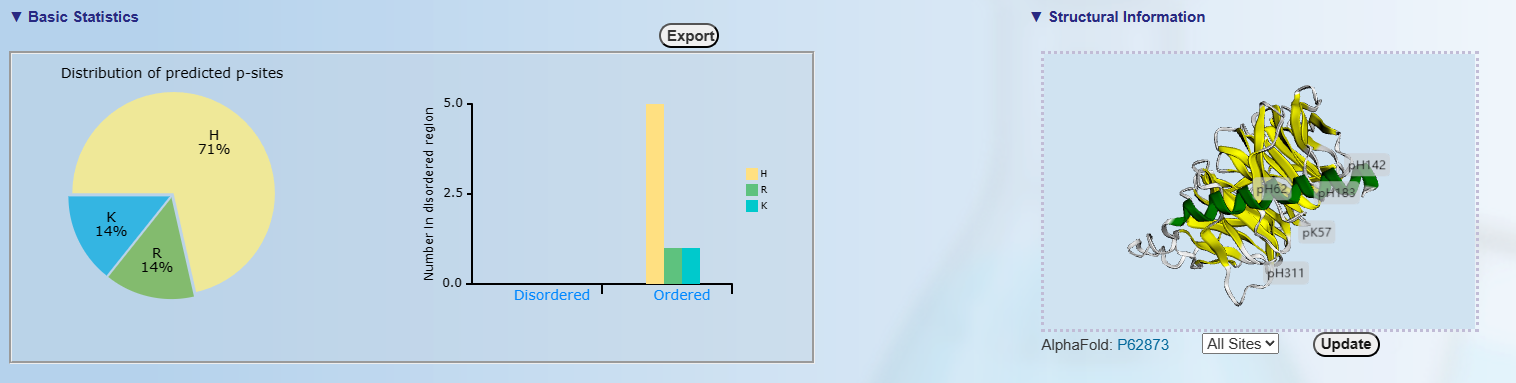
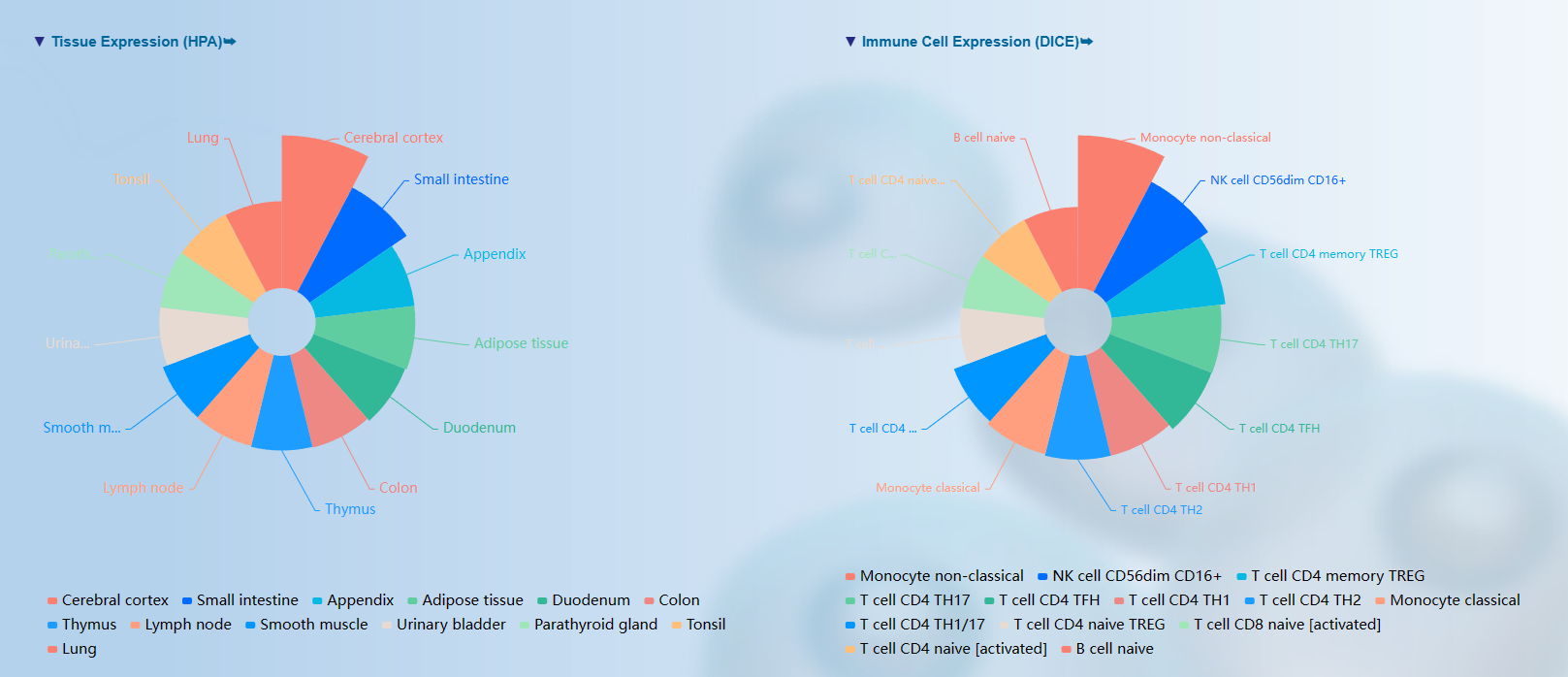
<3>. The visualization of comprehensive prediction
Top: The surface accessibility of amino acids and the protein disordered region were predicted by
NetSurfP ver. 1.1 (PMID: 19646261) and IUPred
(PMID: 15955779), respectively. The cutoff of disordered region prediction=0.5, if score of prediction > cutoff,
the residue is considered in disordered region. The cutoff of surface accessibility prediction=0.25, if score of prediction > cutoff, the residue is considered as surface exposed residue.
Bottom: The positions of the predicted phosphorylation sites were visualized in the protein sequence together with the secondary structure predicted by NetSurfP ver. 1.1 (PMID: 19646261).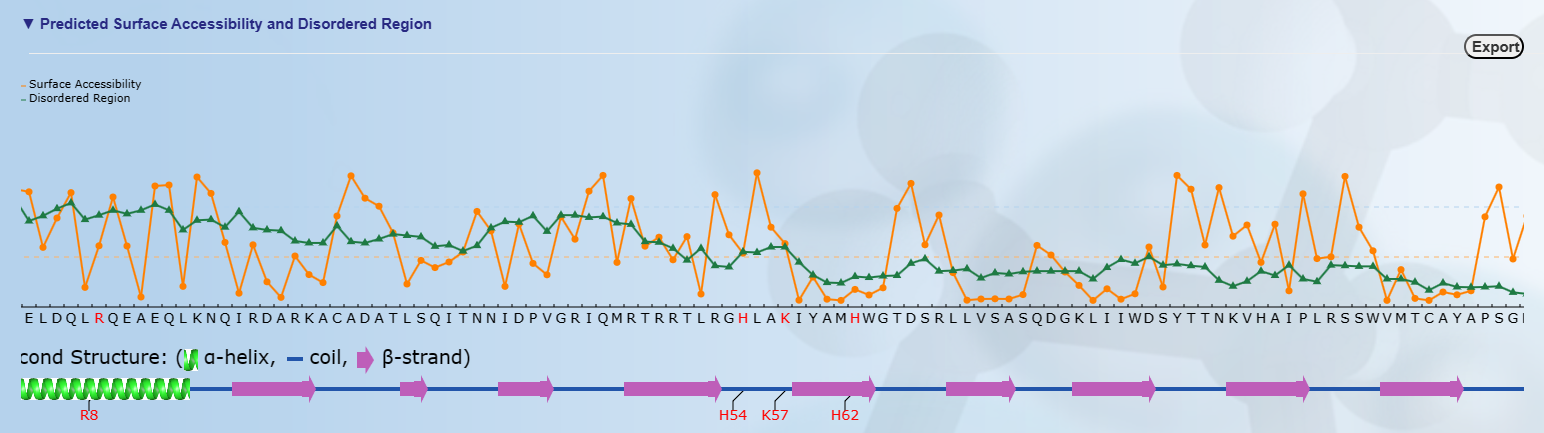
Left: The distribution of H/R/K sites in secondary structure.
Right: The distribution of H/R/K sites in disordered region.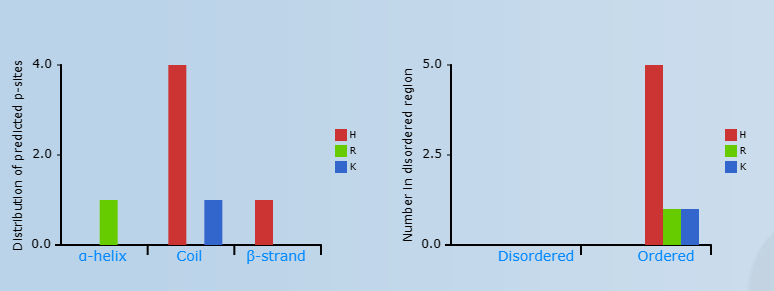
OS Version Chrome Firefox Microsoft Edge Safari Linux Ubuntu 22.04.3 120.0.6099.71 116.0.2 N/A N/A MacOS HighSierra 120.0.6099.62 116.0.3 N/A 17.1 Windows 10 119.0.6045.200 120.0.1 120.0.2210.61 N/A